September 18, 2018
Sharing data sets for collaboration or publication has always been challenging, but it’s become increasingly problematic as complex and high dimensional data sets have become ubiquitous in the life sciences. Studies are large and time consuming; data collection takes time, data analysis is a moving target, as is the software used to carry it out.
In the vaccine space (where I work) we analyze collections of high-dimensional immunological data sets from a variety of different technologies (RNA sequencing, cytometry, multiplexed antibody binding, and others). These data often arise from clinical trials and can involve tens to hundreds of subjects. The data are analyzed by teams of researchers with a diverse variety of goals.
Data from a single study will lead to multiple manuscripts by different principal investigators, dozens of reports, talks, presentations. There are many different consumers of data sets, and results and conclusions must be consistent and reproducible.
Data processing pipelines tend to be study specific. Even though we have great tidyverse tools that make data cleaning easier, every new data set has idiosyncracies and unique features that require some bespoke code to convert them from raw to tidy data.
Best practices tell us raw data are read-only, and analysis is done on some form of processed and tidied data set. Conventional wisdom is that this data tidying takes a substantial amount of the time and effort in a data analysis project, so once it’s done, why should every consumer of a data set repeat it?
One important reason is we don’t always know what a collaborator did to go from raw data to their version of an analysis-ready data set, so the instinct is to ask for the raw data and do the processing ourselves, which involves shipping around inconveniently large files.
 Objectives
Objectives
How can we ensure:
- Data consumers aren’t reinventing the wheel and repeating work already done.
- Data are processed and tidied consistently and reproducibly.
- Everyone is using the same versions of data sets for their analyses.
- Reports and analyses are tied to specific versions of the data and are updated when data change.
 DataPackageR
DataPackageR
Solutions to these issues require reproducible data processing, easy data sharing and versioning of data sets, and a way to verify and track data provenance of raw to tidy data.
Much of this can be accomplished by building R data packages, which are formal R packages whose sole purpose is to contain, access, and / or document data sets. By coopting R’s package system, we get documentation, versioning, testing, and other best practices for free.
DataPackageR was built to help with this packaging process.
 Benefits of DataPackageR
Benefits of DataPackageR
-
It aims to automate away much of the tedium of packaging data sets without getting too much in the way, and keeps your processing workflow reproducible.
-
It sets up the necessary package structure and files for a data package.
-
It allows you to keep the large, raw data sets separate and only ship the packaged tidy data, saving space and time consumers of your data set need to spend downloading and re-processing it.
-
It maintains a reproducible record (vignettes) of the data processing along with the package. Consumers of the data package can verify how the processing was done, increasing confidence in your data.
-
It automates construction of the documentation and maintains a data set version and an md5 fingerprint of each data object in the package. If the data changes and the package is rebuilt, the data version is automatically updated.
-
Data packages can be version controlled on GitHub (or other VCS sites), making for easy change tracking and sharing with collaborators.
-
When a manuscript is submitted based on a specific version of a data package, one can make a GitHub release and automatically push it to sites like zenodo so that it is permanently archived.
-
Consumers of the data package pull analysis-ready data from a central location, ensuring all downstream results rely on consistently processed data sets.
 Development
Development
The package developed organically over the span of several years, initially as a proof-of-concept, with features bolted on over time. Paul Obrecht kindly contributed the data set autodocumentation code and provided in-house testing. As it gained usage internally, we thougt it might be useful to others, and so began the process of releasing it publicly.
 rOpenSci
rOpenSci
DataPackageR was a departure from the usual tools we develop, which live mostly on Bioconductor. We thought the rOpenSci community would be a good place to release the package and reach a wider, more diverse audience.
 The onboarding process
The onboarding process
Onboarding was a great experience. Maëlle Salmon, Kara Woo, Will Landau, and Noam Ross volunteered their time to provide in-depth, comprehensive and careful code review, suggestions for features, documentation, and improvements in coding style and unit tests, that ultimately made the software better. It was an impressive effort (you can see the GitHub issue thread for yourself here).
The current version of the package has 100% test coverage and comprehensive documentation. One great benefit is that as I develop new features in the future, I can be confident I’m not inadvertently breaking something else.
rOpenSci encourages a number of other best practices that I had been somewhat sloppy about enforcing on myself. One is maintaining an updated NEWS.md file which tracks major changes and new features, and links them to relevant GitHub issues. I find this particularly useful as I’ve always appreciated an informative NEWS file to help me decide if I should install the latest version of a piece of software. The online handbook for rOpenSci package development, maintenance and peer review is a great resource to learn about what some of those other best practices are and how you can prepare your own software for submission.
 Using DataPackageR
Using DataPackageR
Here’s a simple example that demonstrates how DataPackageR can be used to construct a data pacakge.
In this example, I’ll tidy the cars data set by giving it more meaningful column names. The original data set has columns speed and dist. We’ll be more verbose and rename them to speed_mph and stopping_distance.
 An outline of the steps involved.
An outline of the steps involved.
- Create a package directory structure. (
DataPackageR::datapackage_skeleton()) - We add a raw data set. (
DataPackageR::use_raw_dataset()) - We add a processing script to process
carsintotidy_cars. (DataPackageR::use_processing_script()) - We define the tidy data object (named
tidy_cars) that will be stored in our data package. (DataPackageR::use_data_object())
 Create a data package directory structure.
Create a data package directory structure.
The first step is to create the data package directory structure. This package structure is based on rrrpkg and some conventions introcuded by Hadley Wickham.
datapackage_skeleton(name = "TidyCars",
path = tempdir(),
force = TRUE)
## ✔ Setting active project to '/private/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T/RtmpXXFG1L/TidyCars'
## ✔ Creating 'R/'
## ✔ Creating 'man/'
## ✔ Writing 'DESCRIPTION'
## ✔ Writing 'NAMESPACE'
## ✔ Added DataVersion string to 'DESCRIPTION'
## ✔ Creating 'data-raw/'
## ✔ Creating 'data/'
## ✔ Creating 'inst/extdata/'
## ✔ configured 'datapackager.yml' file
usethis::proj_set and usethis::proj_get, so be aware if you are using that package in your own scripts, mixing it with DataPackageR, there's a risk of unexpected side-effects.
The directory structure that’s created is shown below.
/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T//RtmpXXFG1L/TidyCars
├── DESCRIPTION
├── R
├── Read-and-delete-me
├── data
├── data-raw
├── datapackager.yml
├── inst
│ └── extdata
└── man
6 directories, 3 files
 Add raw data to the package.
Add raw data to the package.
Next, we work interactively, following the paradigm of the usethis package.
# write our raw data to a csv
write.csv(x = cars,file = file.path(tempdir(),"cars.csv"),row.names = FALSE)
# this works because we called datapackage_skeleton() first.
use_raw_dataset(file.path(tempdir(),"cars.csv"))
use_raw_dataset() moves the file or path in its argument into inst/extdata under the data package source tree. This raw (usually non-tidy) data will be installed with the data pacakge.
For large data sets that you may not want to distribute with the package, you could place them in a directory external to the package source, or place them in inst/extdata but include them in .Rbuildignore. In fact as I write this, there should be an option to add a data set to .Rbuildignore automatically. That would be a good first issue for anyone who would like to contribute.
 Add a data processing script.
Add a data processing script.
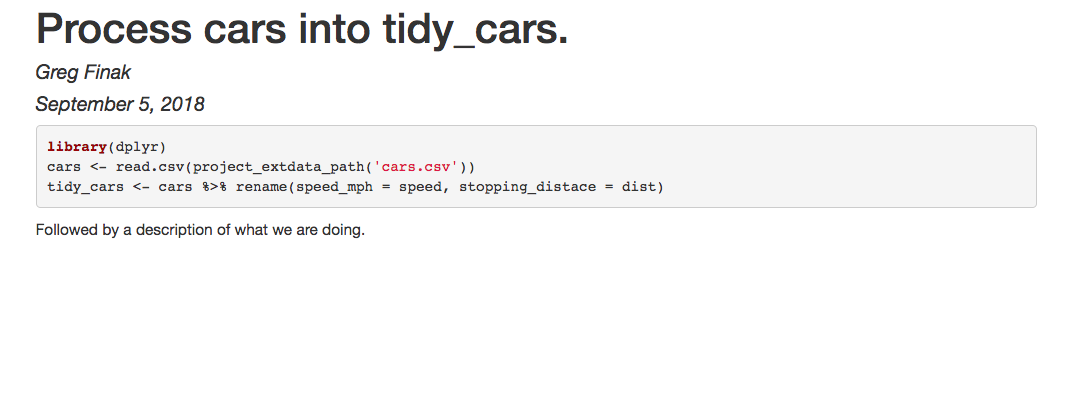
We add a script to process cars into tidy_cars. The author and title are provided as arguments. They will go into the yaml frontmatter of the Rmd file.
Rmd file. This is recommended. Use literate programming to process your data, and the Rmd will appear as a vignette in your final data package.
use_processing_script(file = "tidy_cars.Rmd",
author = "Greg Finak",
title = "Process cars into tidy_cars.")
## configuration:
## files:
## tidy_cars.Rmd:
## enabled: yes
## objects: []
## render_root:
## tmp: '103469'
The script file tidy_cars.Rmd is created in the data-raw directory of the package source. The output echoed after the command is the contents of the datapackager.yml configuration file. It controls the build process. Here the file is added to the configuration. You can find more information about it in the configuration vignette.
 Edit your processing script.
Edit your processing script.
Our script will look like this:
---
title: Process cars into tidy_cars.
author: Greg Finak
date: September 5, 2018
output_format: html_document
---
```{r}
library(dplyr)
cars <- read.csv(project_extdata_path('cars.csv'))
tidy_cars <- cars %>% rename(speed_mph = speed, stopping_distace = dist)
```
Followed by a description of what we are doing.
 An important note about reading raw data from the processing script.
An important note about reading raw data from the processing script.
In order to read raw data sets in a reproducible manner, DataPackageR provides an API call:
project_extdata_path() that returns the absolute path to its argument in inst/extdata in a reproducible way, independent of the current working directory. There are also project_path() and project_data_path() that will point to the source root and the data directory, respectively.
DataPackageR is not compatible with the here package. Rather use the APIs above.This script creates the tidy_cars object, which is what we want to store in our final package.
 Let DataPackageR know about the data objects to store in the package.
Let DataPackageR know about the data objects to store in the package.
We let DataPackageR know about this:
use_data_object("tidy_cars")
## configuration:
## files:
## tidy_cars.Rmd:
## enabled: yes
## objects:
## - tidy_cars
## render_root:
## tmp: '103469'
Again, the datapackager.yml configuration is echoed, and we see the data set object has been added.
The build process uses this file to know which scripts to run and what data outputs to expect. More information is in the technical vignette.
It will automatically create documentation stubs for the package and for these data objects.
 Build the package (for the first time).
Build the package (for the first time).
We build the package. It will automatically generate some documentation for the data sets that we’ll need to go in and edit. There’s also some informative output. The output has been cleaned up recently, particularly now that the package has stabilized.
options("DataPackageR_interact" = FALSE)
package_build() in interactive mode, you'll be prompted to fill in one line of information that will be put in the NEWS.md file, describing the changes to the data package. This helps you track changes across versions.
Setting options("DataPackageR_interact" = FALSE) turns off interactive mode.
package_build(packageName = file.path(tempdir(),"TidyCars"), install = FALSE)
##
## ✔ Setting active project to '/private/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T/RtmpXXFG1L/TidyCars'
## ✔ 1 data set(s) created by tidy_cars.Rmd
## • tidy_cars
## ☘ Built all datasets!
## Non-interactive NEWS.md file update.
##
## ✔ Creating 'vignettes/'
## ✔ Creating 'inst/doc/'
## First time using roxygen2. Upgrading automatically...
## Updating roxygen version in /private/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T/RtmpXXFG1L/TidyCars/DESCRIPTION
## Writing NAMESPACE
## Loading TidyCars
## Writing TidyCars.Rd
## Writing tidy_cars.Rd
## '/Library/Frameworks/R.framework/Resources/bin/R' --no-site-file \
## --no-environ --no-save --no-restore --quiet CMD build \
## '/private/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T/RtmpXXFG1L/TidyCars' \
## --no-resave-data --no-manual --no-build-vignettes
##
## Next Steps
## 1. Update your package documentation.
## - Edit the documentation.R file in the package source data-raw subdirectory and update the roxygen markup.
## - Rebuild the package documentation with document() .
## 2. Add your package to source control.
## - Call git init . in the package source root directory.
## - git add the package files.
## - git commit your new package.
## - Set up a github repository for your pacakge.
## - Add the github repository as a remote of your local package repository.
## - git push your local repository to gitub.
## [1] "/private/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T/RtmpXXFG1L/TidyCars_1.0.tar.gz"
install = FALSE prevents the package from being automatically installed to the system after building.
Above, you’ll see the message:
1 data set(s) created by tidy_cars.Rmd
Indicating that the build process found the one of the expected data sets. It then lists the specific data set(s) it created in that script.
It stores those data sets in the package data subfolder as .rda files.
You’ll see it also created vignettes, wrote a roxygen stub for the documentation of the TidyCars package and the tidy_cars data object. It created provisional Rd files, and built the package archive. It even provides some information on what to do next.
 Next edit the data set documentation.
Next edit the data set documentation.
Let’s update our documentation as requested. We edit the documentation.R file under data-raw.
Here are its contents:
#' TidyCars
#' A data package for TidyCars.
#' @docType package
#' @aliases TidyCars-package
#' @title Package Title
#' @name TidyCars
#' @description A description of the data package
#' @details Use \code{data(package='TidyCars')$results[, 3]} tosee a list of availabledata sets in this data package
#' and/or DataPackageR::load_all
#' _datasets() to load them.
#' @seealso
#' \link{tidy_cars}
NULL
#' Detailed description of the data
#' @name tidy_cars
#' @docType data
#' @title Descriptive data title
#' @format a \code{data.frame} containing the following fields:
#' \describe{
#' \item{speed_mph}{}
#' \item{stopping_distace}{}
#' }
#' @source The data comes from________________________.
#' @seealso
#' \link{TidyCars}
NULL
This is standard roxygen markup. You can use roxygen or markdown style comments. You should describe your data set, where it comes from, the columns of the data (if applicable), and any other information that can help a user make good use of and understand the data set.
We’ll fill this in and save the resulting file.
## #' TidyCars
## #' A data package for TidyCars.
## #' @docType package
## #' @aliases TidyCars-package
## #' @title A tidied cars data set.
## #' @name TidyCars
## #' @description Cars but better. The variable names are more meaninful.
## #' @details The columns have been renamed to indicate the units and better describe what is measured.
## #' @seealso
## #' \link{tidy_cars}
## NULL
##
##
##
## #' The stopping distances of cars at different speeds.
## #' @name tidy_cars
## #' @docType data
## #' @title The stopping distances of cars traveling at different speeds.
## #' @format a \code{data.frame} containing the following fields:
## #' \describe{
## #' \item{speed_mph}{The speed of the vehicle.}
## #' \item{stopping_distace}{The stopping distance of the vehicle.}
## #' }
## #' @source The data comes from the cars data set distributed with R.
## #' @seealso
## #' \link{TidyCars}
## NULL
Then we run the document() method in the DataPackageR:: namespace to rebuild the documentation.
# ensure we run document() from the DataPackageR namespace and not document() from roxygen or devtools.
package_path <- file.path(tempdir(),"TidyCars")
DataPackageR::document(package_path)
##
## ✔ Setting active project to '/private/var/folders/jh/x0h3v3pd4dd497g3gtzsm8500000gn/T/RtmpXXFG1L/TidyCars'
## Updating TidyCars documentation
## Loading TidyCars
## Writing TidyCars.Rd
## Writing tidy_cars.Rd
## [1] TRUE
 Iterate…
Iterate…
We can add more data sets, more scripts, and so forth, until we’re happy with the package.
 A final build.
A final build.
Finally we rebuild the package one last time. The output is suppressed here for brevity.
package_build(file.path(tempdir(),"TidyCars"))
 Sharing and distributing data packages.
Sharing and distributing data packages.
If you are the data package developer you may consider:
- Placing the source of the data package under version control (we like git and GitHub).
- Share the package archive (
yourpackage-x.y.z.tar.gz)- on a public repository.
- directly with collaborators.
We’ve placed the TidyCars data package on GitHub so that you can see for yourself how it looks.
 Limitations and future work
Limitations and future work
Versions of software dependencies for processing scripts are not tracked. Users should use sessionInfo() to keep track of the versions of software used to perform data processing so that the environment can be replicated if a package needs to be rebuilt far in the future.
Tools like packrat and Docker aim to solve these problems, and it is non-trivial. I would love to integrate these tools more closely with DataPackageR in the future.
 Using a data package.
Using a data package.
If you are a user of a data pacakge.
Install a data package in the usual manner of R package installation.
- From GitHub:
devtools::install_github("gfinak/TidyCars")
- From the command line:
R CMD INSTALL TidyCars-0.1.0.tar.gz
Once the package is installed, we can load the newly built package and access documentation, vignettes, and use the DataVersion of the package in downstream analysis scripts.
library(TidyCars)
browseVignettes(package = "TidyCars")
Typing the above in the R console will pop up a browser window where you’ll see the available vignettes in your new TidyCars package.

Clicking the HTML link gives you access to the output of the processing script, rendered as a vignette. Careful work here will let you come back to your code and understand what you have done.
We can also view the data set documentation:
?tidy_cars

And we can use the assert_data_version() API to test the version of a data package in a downstream analysis that depends on the data.
data_version("TidyCars")
## [1] '0.1.0'
assert_data_version("TidyCars",version_string = "0.1.0", acceptable = "equal")
assert_data_version("TidyCars",version_string = "0.2.0", acceptable = "equal")
## Error in assert_data_version("TidyCars", version_string = "0.2.0", acceptable = "equal"): Found TidyCars 0.1.0 but == 0.2.0 is required.
The first assertion is true, the second is not, throwing an error. In downstream analyses that depend on a version of a data package, this is useful to ensure updated data don’t inadvertently change results, without the user being aware that something unexpected is going on.
A data package can be built into a package archive (.tar.gz) using the standard R CMD build process. The only difference is that the .rda files in /data won’t be re-generated, and the existing vignettes describing the processing won’t be rebuilt. This is useful when the processing of raw data sets is time consuming (like some biological data), or when raw data sets are too large to distribute conveniently.
 To conclude
To conclude
With DataPackageR I’ve tried to implement a straightforward workflow for building data packages. One that doesn’t get in the way (based on my own experience) of how people develop their data processing pipelines. The new APIs are limited and only minor changes need to be made to adapt existing code to the workflow.
With a data package in hand, data can be easily shared and distributed. In my view, the greatest benefit of building a data package is that it encourages us to use best practices, like documenting data sets, documenting code, writing unit tests, and using version control. Since we are very often our own worst enemies, these are all Good Things.
We have been eating our own dog food, so to speak. We use data packages internally in the Vaccine Immunology Statistical Center to prepare data sets for analysis, publication, and for sharing with collaborators. We often do so through the Fred Hutch GitHub Organization (though most data are private).
The RGLab has used data packages (though not built using DataPackageR) to share data sets together with publications:
- Combinatorial Polyfunctionality Analysis of Single Cells: paper, data
- Model-based Analysis of Single-cell Trascriptomics paper, data
In the long run, packaging data saves us time and effort (of the sort expended trying to reproduce results of long ago and far away) by ensuring data processing is reproducible and properly tracked. It’s proved extremely useful to our group and I hope it’s useful to others as well.

